
However, there is a tradeoff between microscope availability, high-throughput capacity and image quality, often forcing researchers to use low-end stereoscopic microscopes that can suffice for qualitative assessment. Generally, high-content screening (HCS) devices represent an ideal platform to image a large number of samples under a variety of conditions ( Durens et al., 2020 Brandenberg et al., 2020 Vrij et al., 2016). Another hindrance is the limited access to an imaging system that can accommodate the variety of plates and devices used in organoid culture ( Rossi et al., 2018). When quantitatively interpreted, such data have been crucial in dissecting the mechanisms responsible for organoid development ( Phipson et al., 2019 Lukonin et al., 2020 Hof et al., 2021). There is therefore now a major bottleneck in the ability to inspect this huge number of images and quantify morphological and fluorescence parameters in space and time with high accuracy and in an unbiased manner. In recent years, due to novel engineering solutions and the need of buffering the large variability of organoid generation ( Gritti et al., 2021), the number of experimental conditions have grown combinatorially and it is now possible to generate increasingly large datasets.
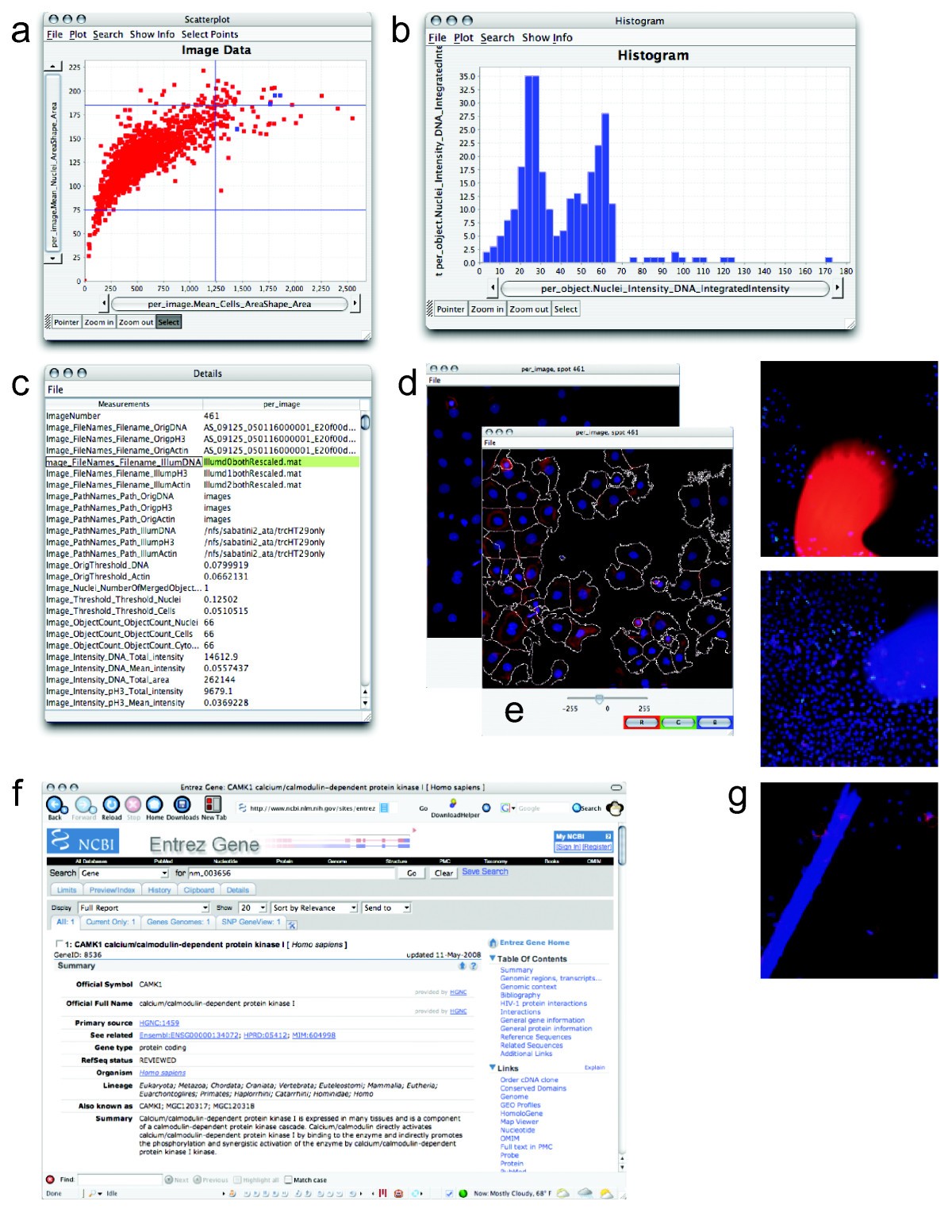
#Cellprofiler analysing a set of images software
We showcase the versatility of MOrgAna on several in vitro systems, each imaged with a different microscope, thus demonstrating the wide applicability of the software to diverse organoid types and biomedical studies. Although the MOrgAna interface is developed for users with little to no programming experience, its modular structure makes it a customizable package for advanced users. Here, we present MOrgAna, a Python-based software that implements machine learning to segment images, quantify and visualize morphological and fluorescence information of organoids across hundreds of images, each with one object, within minutes. Hence, there is a pressing demand for a coding-free, intuitive and scalable solution that analyses such image data in an automated yet rapid manner. The large volumes of images, resulting from hundreds of organoids cultured at once, are becoming increasingly difficult to inspect and interpret. Organoids are large structures with high phenotypic complexity and are imaged on a wide range of platforms, from simple benchtop stereoscopes to high-content confocal-based imaging systems. In the work of protein subcellular localization, our method provides a framework for processing and classifying microscope images, and further lays the foundation for the study of protein and gene functions.Recent years have seen a dramatic increase in the application of organoids to developmental biology, biomedical and translational studies. Experiments show the validity and effectiveness of our method on yeast cell images, it can significantly improve the accuracy of high-throughput microscopy image-based protein subcellular localization, and we achieve the classification accuracy of 0.9098 on the high-throughput microscopy images of yeast cells. What is more, we use an ensemble method to fuse the classification results from the multi-model structure to obtain the final subcellular location of each single-cell image. In addition, we add Squeeze-and-Excitation Blocks to the network to emphasize more informative features. First of all, we employ a deep convolutional neural network as multi-scale feature extractor and use global average pooling to map extracted features at different stages into feature vectors, then concatenate these multi-scale features to form a multi-model structure for image classification. So in this study, we propose a multi-scale multi-model deep neural network via ensemble strategy for protein subcellular localization on single-cell high-throughput images. However, in the existing image-based protein subcellular localization methods, the traditional techniques are lack of efficiency and accuracy, and the potential of deep learning methods has not been fully tapped. A large number of single-cell high-throughput microscopy images provide us with relevant resources for studying protein distribution patterns. By detecting the abnormalities in subcellular locations, we can infer the occurrence of some diseases and mine new drug targets. Protein subcellular locations are closely related to the function of proteins.


 0 kommentar(er)
0 kommentar(er)
